Simulation of a Stochastic Model for the AKAR4 reaction network
simAKAR4stochastic.Rmd
require(SBtabVFGEN)
#> Loading required package: SBtabVFGEN
library(uqsa)
library(GillespieSSA2)This article provides code to simulate the AKAR4 stochastic model (one time, no sampling).
The Stochastic Model
When the copy number of molecular species in a reaction network system (AKAR4 in our case) is low, we cannot model the amount of molecules deterministically (e.g., with an ODE model), because the stochasticity in the reactions that take place cannot be ignored. In particular, the time at which reactions take place is random, as well as the specific reactions that take place (i.e., what pair of molecules react). To model such system we can use the master equation: we model the (integer) number of each molecule species in the system (e.g., proteins) and how the number of each molecule type (randomly) evolves in time. We can easily sample trajectories from this stochastic model using the Gillespie’s Stochastic Simulation algorithm. Given the current amount of each molecular species in the system at a given time point, we can sample the time at which the next reaction takes place and we can sample the type of reaction (i.e., what pair of molecules react).
To obtain the stochastic model for the reaction network, we need to determine the reaction propensities. These can be derived from the reaction rate coefficients (which are the parameters that we usually use in our models and on which we perform uncertainty quantification). To derive the reaction propensities we referred to this article.
Load the Model
The AKAR4 model is included with the package. To load your own model, see the article “Build and simulate your own Model” article.
# Load the files with information on AKAR4 model and corresponding experimental data
modelFiles <- uqsa_example("AKAR4",full.names=TRUE)
# Save the information from the files into the R variable 'SBtab'
SBtab <- SBtabVFGEN::sbtab_from_tsv(modelFiles)
#> [tsv] file[1] «AKAR4_100nM.tsv» belongs to Document «AKAR4»
#> I'll take this as the Model Name.
#> AKAR4_100nM.tsv AKAR4_25nM.tsv AKAR4_400nM.tsv AKAR4_Compound.tsv AKAR4_Experiments.tsv AKAR4_Output.tsv AKAR4_Parameter.tsv AKAR4_Reaction.tsv
# load existing functions related to the model, e.g., AKAR4_default() parameters
source(uqsa_example("AKAR4",pat='^AKAR4[.]R$'))
print(AKAR4_default())
#> kf_C_AKAR4 kb_C_AKAR4 kcat_AKARp
#> 0.018 0.106 10.200Load Experiments (data)
experiments <- sbtab.data(SBtab)
# for example, these is the initial state of experiment 1:
print(experiments[[1]]$initialState)
#> AKAR4 AKAR4_C AKAR4p C
#> 0.2 0.0 0.0 0.4
# pick parameters for simulation
p <- SBtabVFGEN::sbtab_quantity(SBtab$Parameter)
print(p)
#> kf_C_AKAR4 kb_C_AKAR4 kcat_AKARp
#> 0.018 0.106 10.200Simulate
Function simulator.stoch will output a function
s, which will always simulate the scenarios described in
experiment e (i.e., same initial conditions, same inputs),
but for user supplied parameters.
# generate a function (s) that simulates a trajectory given a parameter in input
ssa2 <- simulator.stoch(experiments[1], model.tab = SBtab, vol = 4e-16, nStochSim = 3, outputFunction=AKAR4_func)
# vol indicates the volume in m^3 where the reactions take place
# nStochSim indicates how many times we simulate the stochastic model; the output trajectory is the average of all (in this case nStochSim = 3) the simulated trajectories
# simulate a trajectory (y) given parameter p
y_ssa2 <- ssa2(p)Plot
par(bty='n',xaxp=c(80,120,4))
plot(
experiments[[1]]$outputTimes,
experiments[[1]]$outputValues$AKAR4pOUT,
ylim=c(90,200), ylab="AKAR4p", xlab="time",
main=names(experiments)[1],
lwd=1.5)
lines(experiments[[1]]$outputTimes, as.numeric(y_ssa2[[1]]$output), col="blue")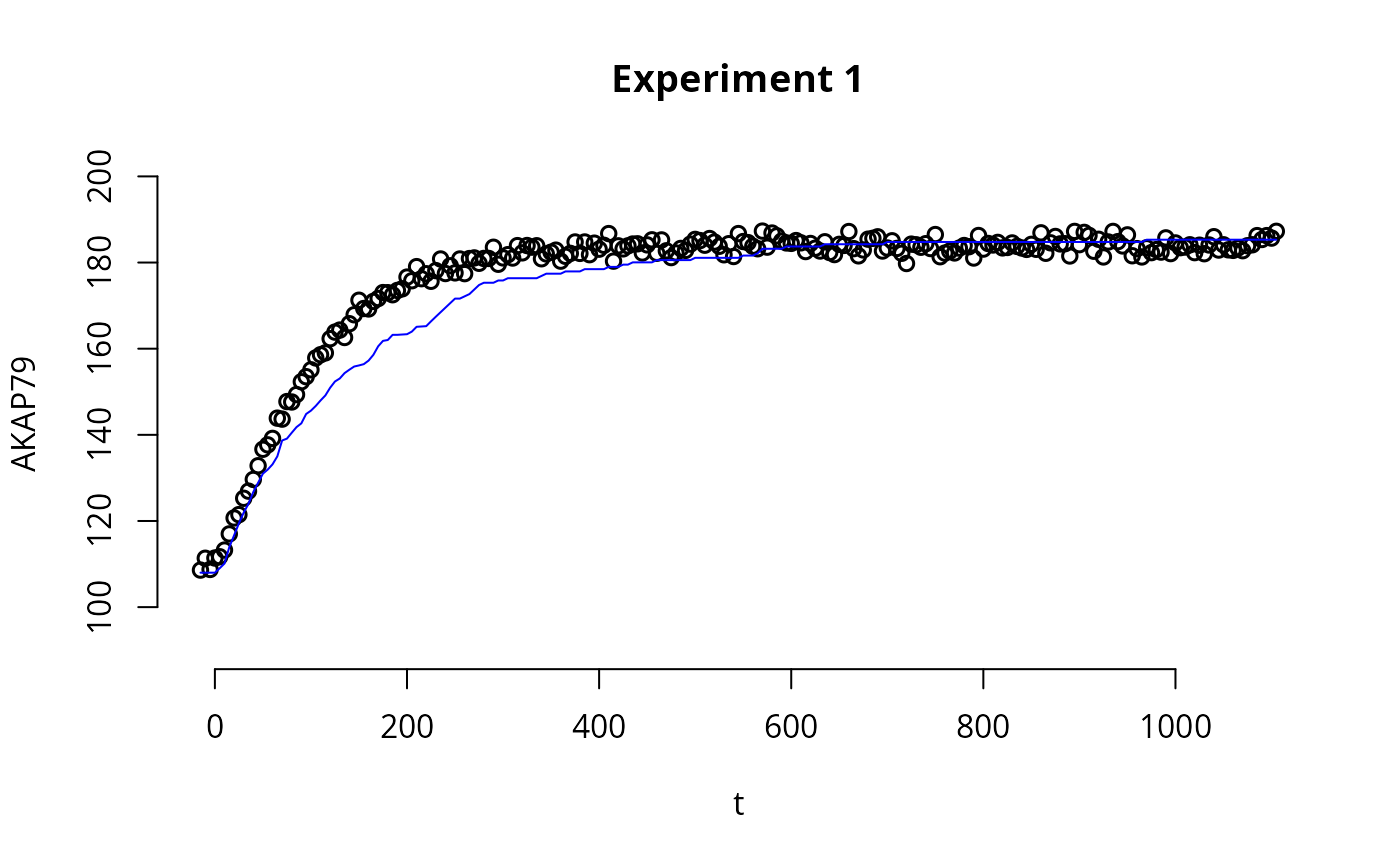
Show Results via ggplot2
if (require(ggplot2)){
D<-data.frame(time=experiments[[1]]$outputTime,
AKAR4p=experiments[[1]]$outputValues$AKAR4pOUT,
AKAR4pERR=experiments[[1]]$errorValues$AKAR4pOUT,
sim=y_ssa2[[1]]$output)
ggplot(D) +
geom_linerange(mapping=aes(x=time,y=AKAR4p,ymin=AKAR4p-AKAR4pERR,ymax=AKAR4p+AKAR4pERR),na.rm=TRUE) +
geom_point(mapping=aes(x=time,y=AKAR4p),na.rm=TRUE) +
geom_line(mapping=aes(x=time,y=sim),color="purple",lwd=1.2)
}
#> Loading required package: ggplot2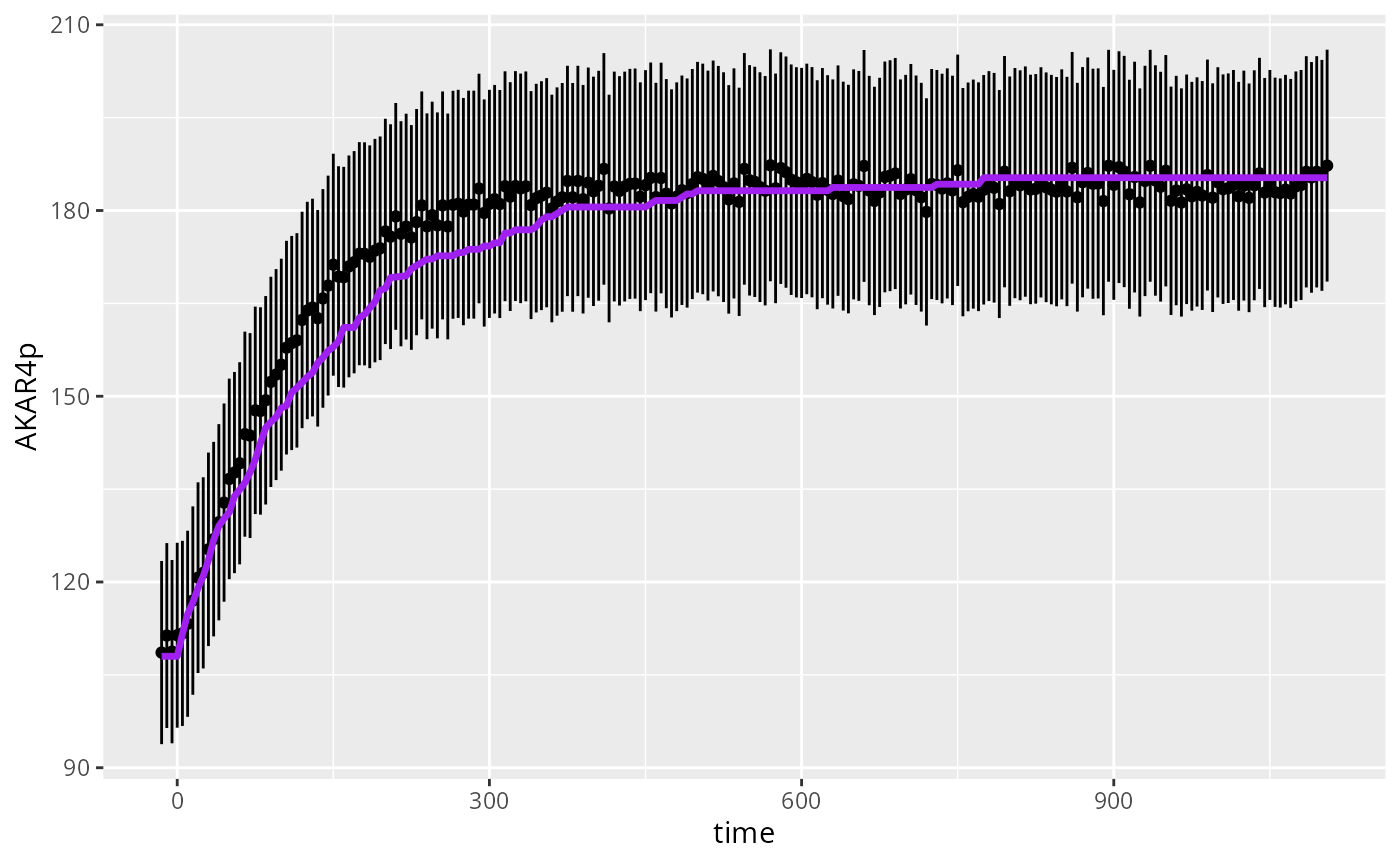
Built-in simulator
Another way to simulate a stochastic model is through the
uqsa own built-in simulator. This assumes that the model in
question comes from an SBtab file and is formulated with concentrations
and kinetic laws.
We auto-generate C code, which uses the kinetic laws, but rescales
all parameters internally to have the unit 1/s and all
compound species are rescaled to be in particle counts. So, the internal
simulator can be used with the list of experiments that we use for ODE
models, with no changes. This rescaling is performed as described in this article.
C <- generateGillespieCode(SBtab)
cat(C,sep="\n",file="AKAR4_st.c")We compile that model to a shared library:
And next we simulate this compiled model:
st <- uqsa::simstoch(experiments,model.so="./akar4-st.so")
#> Warning in uqsa::simstoch(experiments, model.so = "./akar4-st.so"): experiments
#> have no input parameters, defaults to numeric(0).
#> Warning in uqsa::simstoch(experiments, model.so = "./akar4-st.so"): experiments
#> have no input parameters, defaults to numeric(0).
#> Warning in uqsa::simstoch(experiments, model.so = "./akar4-st.so"): experiments
#> have no input parameters, defaults to numeric(0).
y <- st(p)And also plot the results:
tm <- experiments[[1]]$outputTimes
ov <- experiments[[1]]$outputValues$AKAR4pOUT
ev <- experiments[[1]]$errorValues$AKAR4pOUT
par(bty="n")
plot(tm,ov,xlab="time",ylab="AKAR4p",main=names(experiments)[1],ylim=c(90,210))
arrows(tm,ov,tm,ov+ev,angle=90,length=0.02)
arrows(tm,ov,tm,ov-ev,angle=90,length=0.02)
lines(tm,y[[1]]$func["AKAR4pOUT",,],lwd=2, col="red")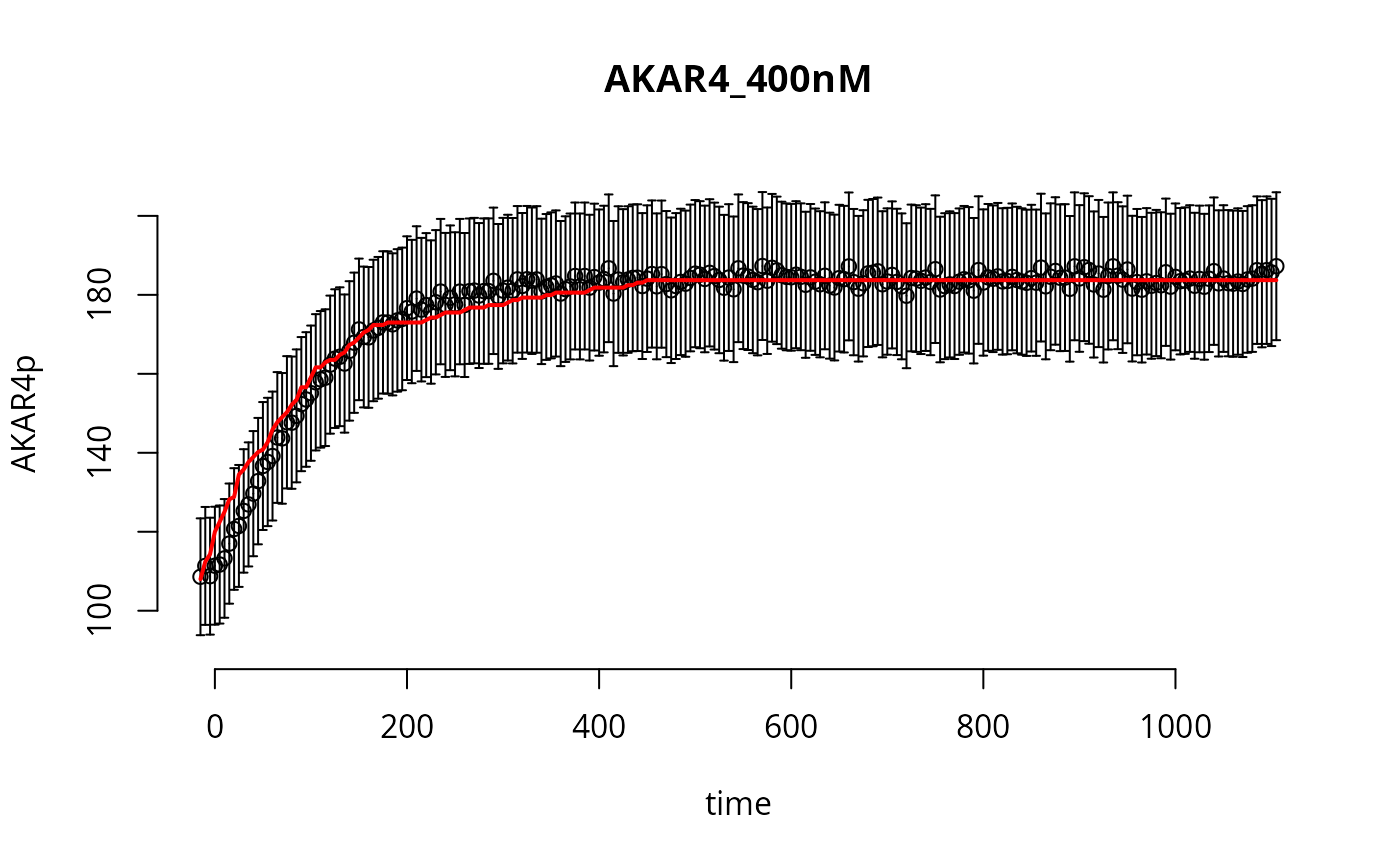
Benchmark
Here we test which of the two simulates faster:
if (require(rbenchmark)){
BM <- benchmark(
GillespieSSA2 = {y1<-ssa2(p)},
uqsa = {y2 <- st(p)},
replications = 500
)
}
#> Loading required package: rbenchmark
print(BM)
#> test replications elapsed relative user.self sys.self user.child
#> 1 GillespieSSA2 500 7.027 106.47 6.995 0.032 0
#> 2 uqsa 500 0.066 1.00 0.066 0.000 0
#> sys.child
#> 1 0
#> 2 0