Uncertainty Quantification of the AKAR4 (stochastic model)
sampleAKAR4stochastic.RmdThis article provides code to perform uncertainty quantification (UQ) of a stochastic reaction network model. As an example we use the AKAR4 stochastic model. The UQ method that we use is ABCMCMC.
For information about the stochastic model, please read the article Simulate AKAR4 stochastically.
Load the Model
This model is included with the package. To load your own model, see the article “Build and simulate your own Model”.
modelFiles <- uqsa_example("AKAR4",full.names=TRUE)
SBtab <- SBtabVFGEN::sbtab_from_tsv(modelFiles)
#> [tsv] file[1] «AKAR4_100nM.tsv» belongs to Document «AKAR4»
#> I'll take this as the Model Name.
#> AKAR4_100nM.tsv AKAR4_25nM.tsv AKAR4_400nM.tsv AKAR4_Compound.tsv AKAR4_Experiments.tsv AKAR4_Output.tsv AKAR4_Parameter.tsv AKAR4_Reaction.tsv
# model related functions, in R, e.g. AKAR4_default() parameters
source(uqsa_example("AKAR4",pat='^AKAR4[.]R$'))
print(AKAR4_default())
#> kf_C_AKAR4 kb_C_AKAR4 kcat_AKARp
#> 0.018 0.106 10.200To simulate the stochastic reaction network model we use the
Gillespie algorithm. Specifically, we use the functions
implemented in the package GillespieSSA2. To use this package we need to
create a variable reactions using the uqsa function
importReactionsSSA with the SBtab model as input.
# Create reactions to simulate the stochastic model with the GillespieSSA2 package
reactions <- importReactionsSSA(SBtab)Moreover, to simulate the stochastic reactions with the Gillespie
algorithm we need to specify the volume of space where the reactions
take place and obtain the parameter Phi which will be
needed for simulations. See this article for more
information
AvoNum <- 6.022e23 #Avogadro constant
unit <- 1e-6 # unit of measure with respect to m^3 (1e-6 corresponds to micrometers)
vol <- 4e-17 # volume where the reactions take place
Phi <- AvoNum * vol * unit # parameter used in the simulationsLoad Experiments (data)
experiments <- sbtab.data(SBtab)
# for example, these is the initial state of experiment 1:
print(experiments[[1]]$initialState)
#> AKAR4 AKAR4_C AKAR4p C
#> 0.2 0.0 0.0 0.4
# save parameters' names
par_val <- SBtab$Parameter[["!DefaultValue"]]
par_names <- SBtab$Parameter[["!Name"]]
names(par_val) <- par_names
print(par_val)
#> kf_C_AKAR4 kb_C_AKAR4 kcat_AKARp
#> 0.018 0.106 10.200Define MCMC settings
# a function that tansforms the ABC variables to acceptable model
# parameters, re-indexing could also happen here
parMap <- function (parABC=0) {
return(10^parABC)
}
# scale to determine prior values
defRange <- 1000
# Define Lower and Upper Limits for logUniform prior distribution for the parameters
ll <- c(par_val/defRange)
ul <- c(par_val*defRange)
ll = log10(ll) # log10-scale
ul = log10(ul) # log10-scale
# Define the prior distribution. In this case Uniform(ll,ul)
dprior <- dUniformPrior(ll, ul) # dprior evaluates the prior density function
rprior <- rUniformPrior(ll, ul) # rprior generates random samples from the prior
# Define Number of Samples for the Precalibration (npc) and each ABC-MCMC chain (ns)
nChains <- 4 # number of ABCMCMC chains to run
ns <- 1000 # no of samples required from each ABC-MCMC chain
npc <- 100 # pre-calibration
# Function to compute the score (distance) between experimental and simulated data
distance <- function(funcSim, dataExpr, dataErr = 1.0){
return(mean(((funcSim-dataExpr)/max(dataExpr))^2,na.rm=TRUE))
}
# Threshold in the ABCMCMC algorithm
delta <- 0.0005Generate an objective function
An objective function in this case is a function that can
read the experimental conditions (initial state, inputs etc.) and the
experimental data in a list of experiments (variable
experiments in the code). Give a parameter in input, for
each experimental condition specified in the list of experiments, the
objective function: 1. simulates the stochastic model given
such parameter, and 2. computes the distance between the simulated
trajectories and the observed trajectories (experimental data).
To generate the objective function for stochastic models you can use
the uqsa function makeObjectiveSSA.
objectiveFunction <- makeObjectiveSSA(experiments = experiments, model.tab = SBtab, parNames = par_names, outputFunction = model$func, distance = distance, parMap = parMap, reactions = reactions, nStochSim = 3)Run PreCalibration sampling
Before running the ABCMCMC code, we run a precalibration to determine good initial values and algorithmic hyperparameters for the ABCMCMC chains.
p <- 0.01 # Choose Top 1% samples from the precalibration samples with shortest distance to the experimental values
sfactor <- 0.1 # Scaling factor used to determine a good covariance matrix for the ABCMCMC proposal moves
pC <- preCalibration(objectiveFunction, npc, rprior, rep = 1, p=p, sfactor=sfactor, delta=delta, num=nChains)
#> Warning in getMCMCPar(prePar, preDelta, p = p, sfactor = sfactor, delta = delta, : distances between experiment and simulation are too big; selecting the best (112) parameter vectors.
Sigma <- pC$Sigma
startPar <- pC$startPar
for(j in 1 : nChains){
cat("Chain", j, "\n")
cat("\tMin distance of starting parameter for chain",j," = ", min(objectiveFunction(startPar[,j])),"\n")
cat("\tMean distance of starting parameter for chain",j," = ", mean(objectiveFunction(startPar[,j])),"\n")
cat("\tMax distance of starting parameter for chain",j," = ", max(objectiveFunction(startPar[,j])),"\n")
}
#> Chain 1
#> Min distance of starting parameter for chain 1 = 0.004448397
#> Mean distance of starting parameter for chain 1 = 0.05343058
#> Max distance of starting parameter for chain 1 = 0.1265913
#> Chain 2
#> Min distance of starting parameter for chain 2 = 0.003646814
#> Mean distance of starting parameter for chain 2 = 0.05872865
#> Max distance of starting parameter for chain 2 = 0.1402112
#> Chain 3
#> Min distance of starting parameter for chain 3 = 0.0001839402
#> Mean distance of starting parameter for chain 3 = 0.002454254
#> Max distance of starting parameter for chain 3 = 0.004795667
#> Chain 4
#> Min distance of starting parameter for chain 4 = NaN
#> Mean distance of starting parameter for chain 4 = NaN
#> Max distance of starting parameter for chain 4 = NaNRun ABC-MCMC Sampling
The sampling is parallelized on several cores using the
parLapply function. Running this code will take
approximately 10 minutes on a 8-core laptop.
cl <- parallel::makeForkCluster(nChains, outfile="outputMessagesABCMCMC.txt")
out_ABCMCMC <- parLapply(
cl,
1:nChains,
function(j) {
tryCatch(
ABCMCMC(
objectiveFunction, startPar[,j], ns, Sigma, delta, dprior, batchSize = 10),
error=function(cond) {message("ABCMCMC crashed"); return(NULL)}
)
}
)
stopCluster(cl)
ABCMCMCoutput <- do.call(Map, c(rbind,out_ABCMCMC))
colnames(ABCMCMCoutput$draws) <- par_namesVisualize the sampled parameters
The parameters in the variable ABCMCMCoutput are
approximate samples from the posterior distribution of the parameters
given the considered experiments.
The sampled three dimensional parameters for the AKAR4 model can be visualized via scatter plots.
if (require(hexbin)){
hexbin::hexplom(ABCMCMCoutput$draws, xbins = 16)
} else {
pairs(ABCMCMCoutput$draws, pch=20)
}
#> Loading required package: hexbin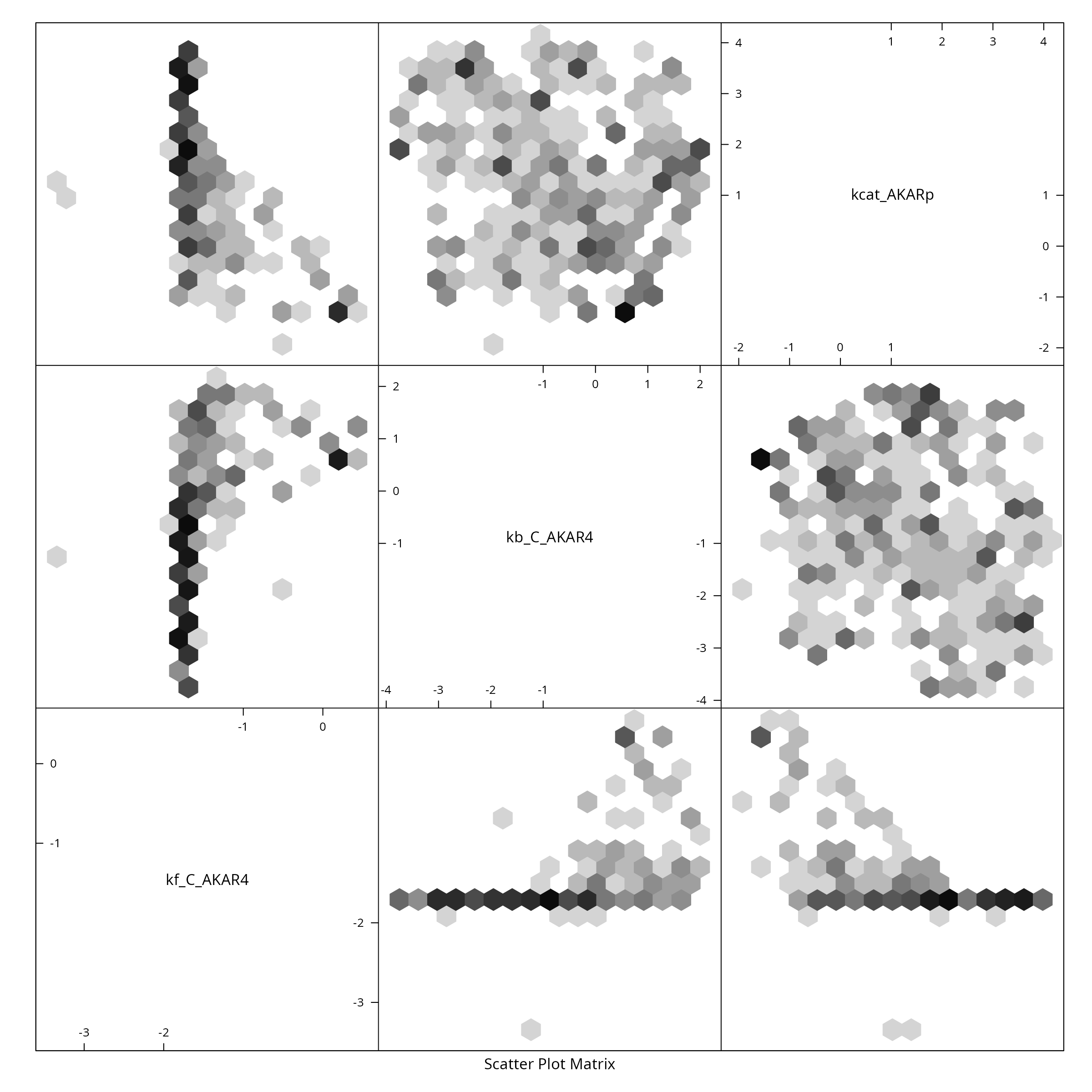
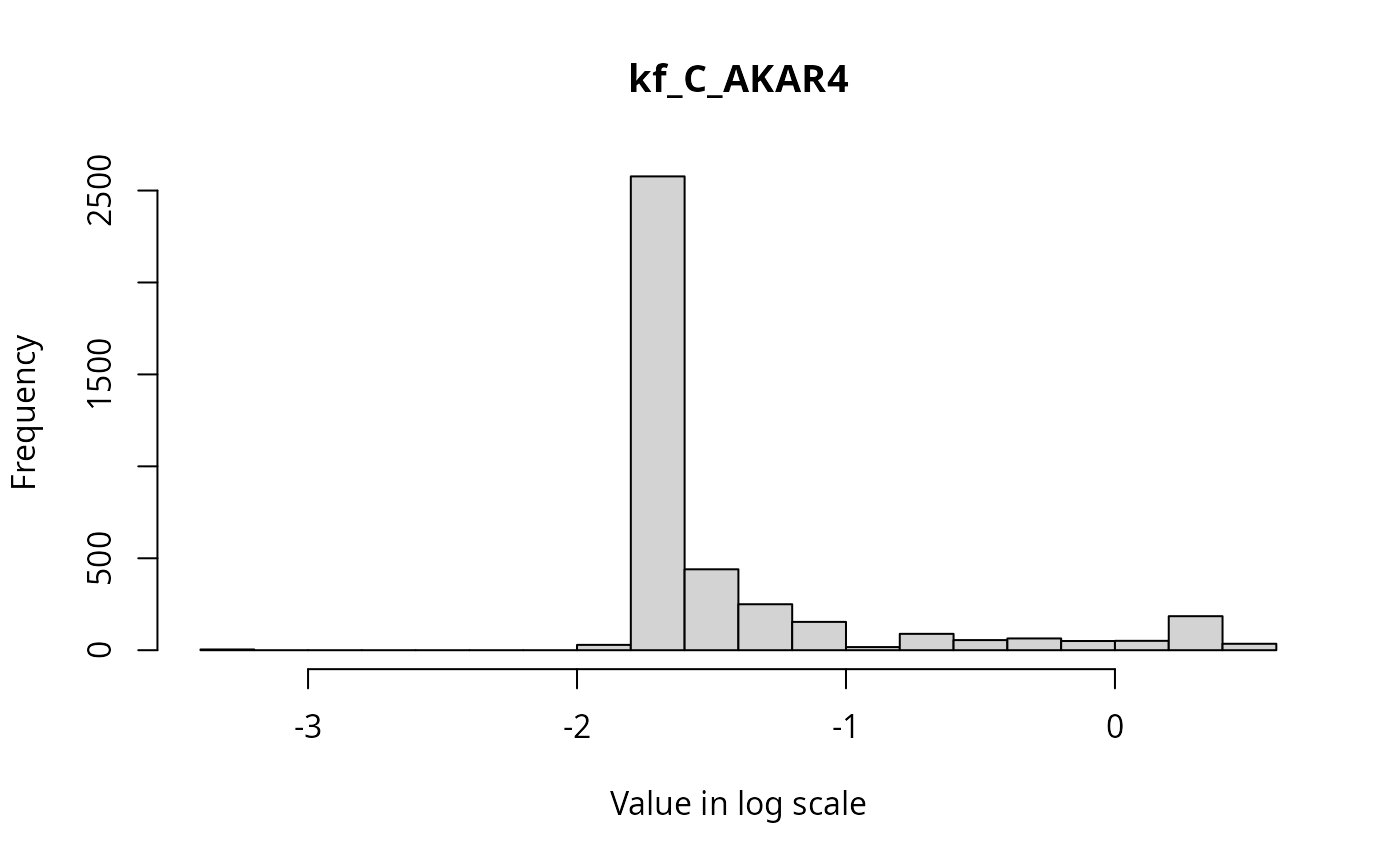
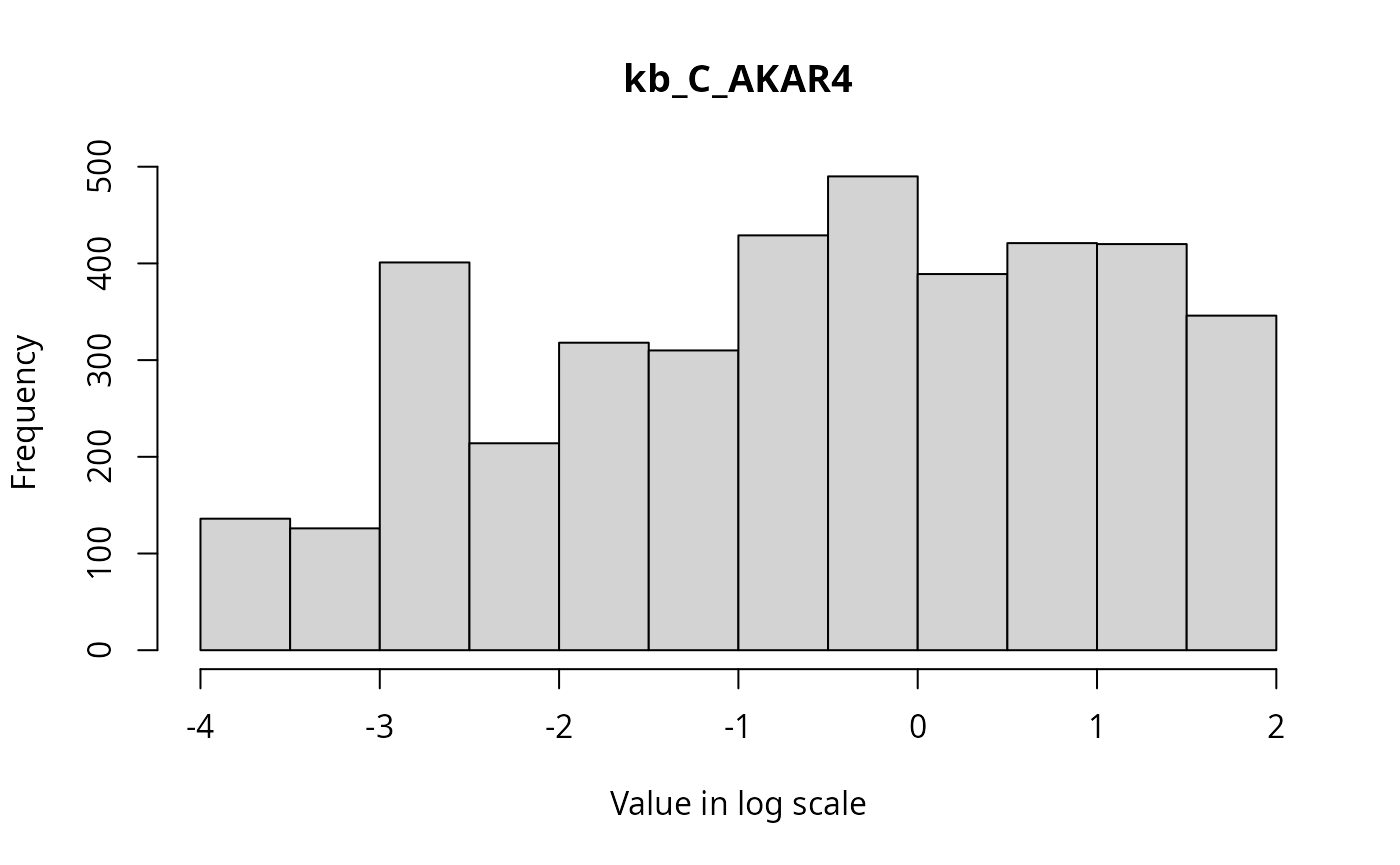
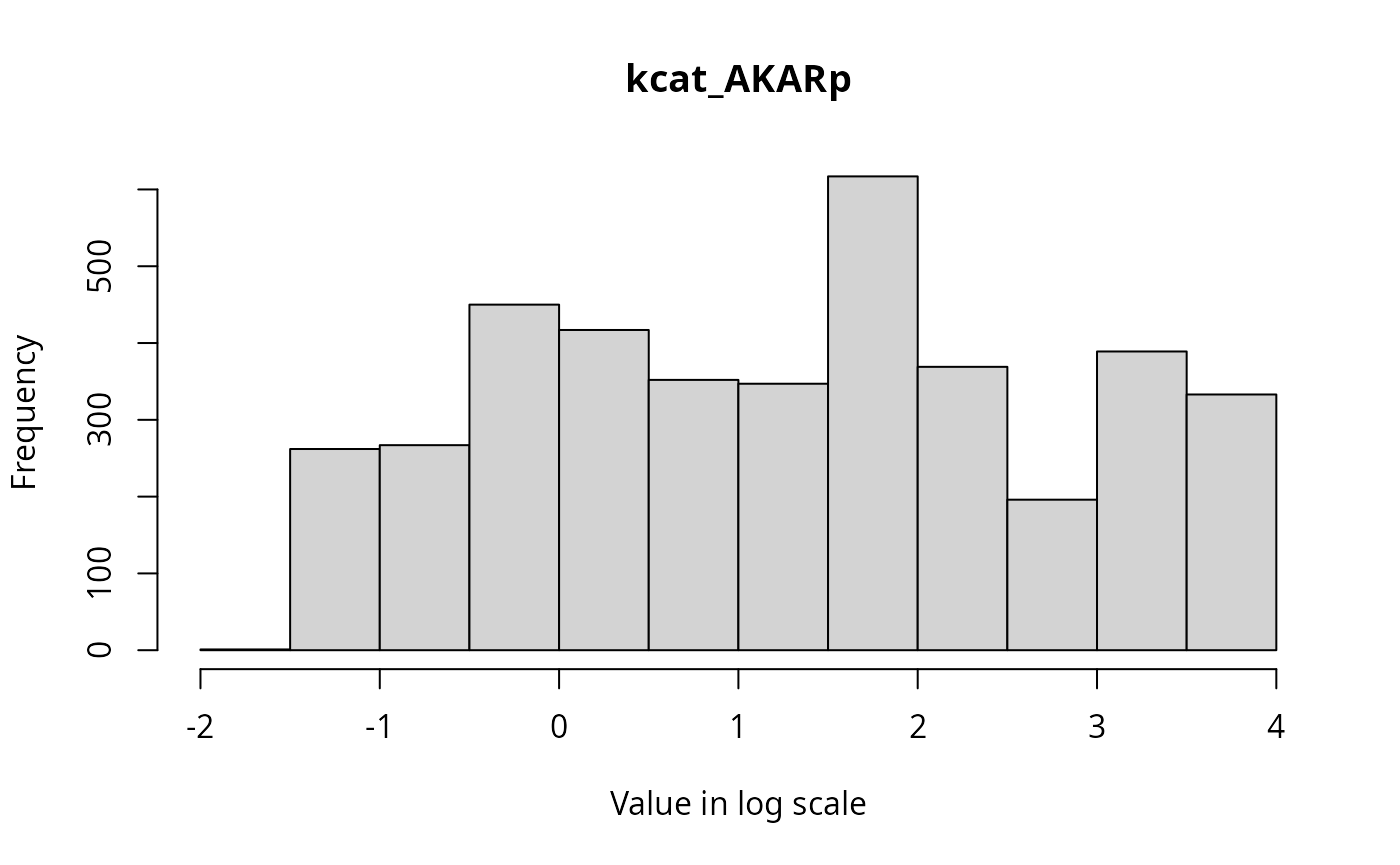
The marginal distributions of each parameter in the parameter vector can be visualized via histograms as follows.
for(i in 1:3){
hist(ABCMCMCoutput$draws[,i], breaks = 20, main=par_names[i], xlab = "Value in log scale")
}
Simulate the stochastic model given posterior samples
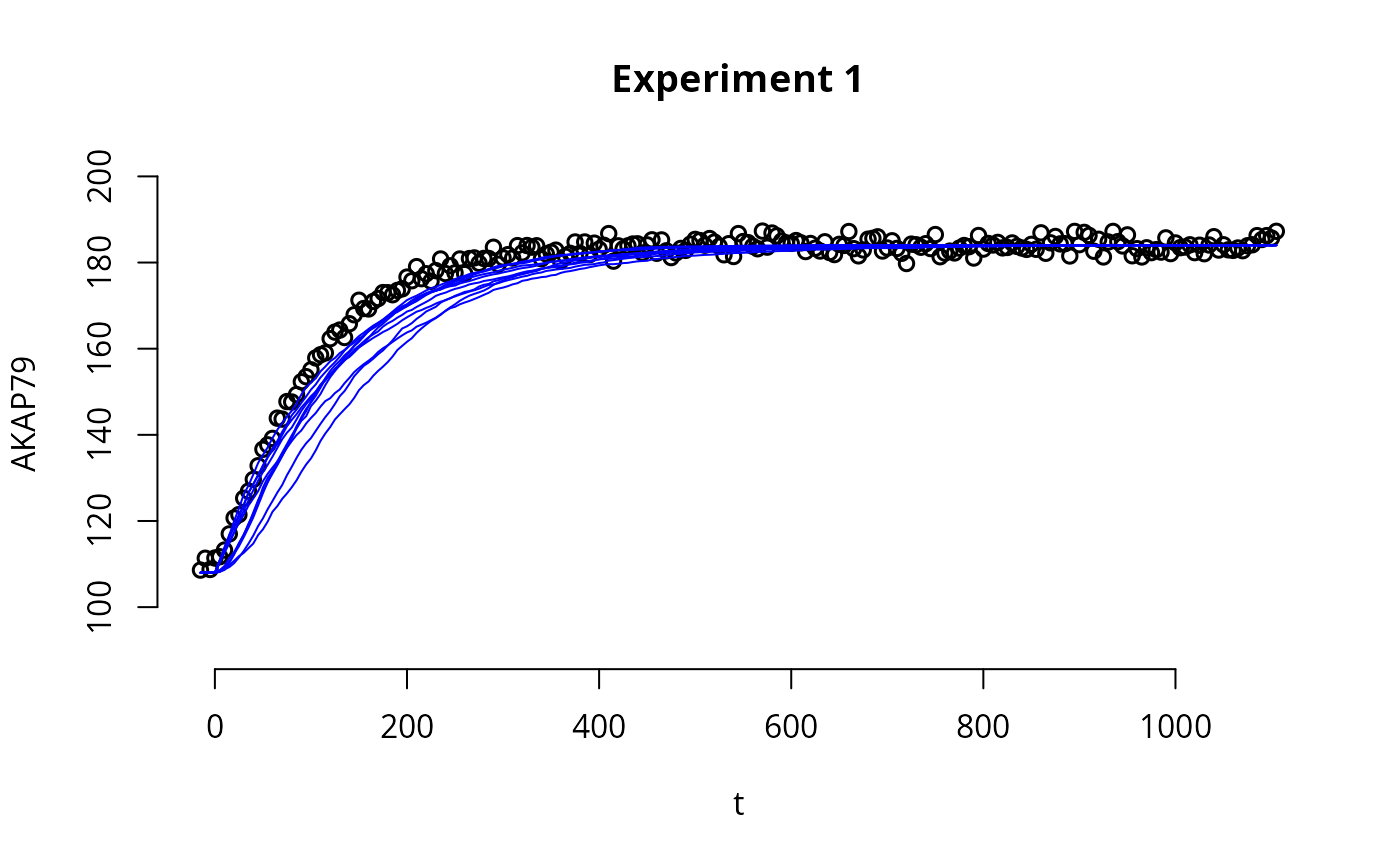
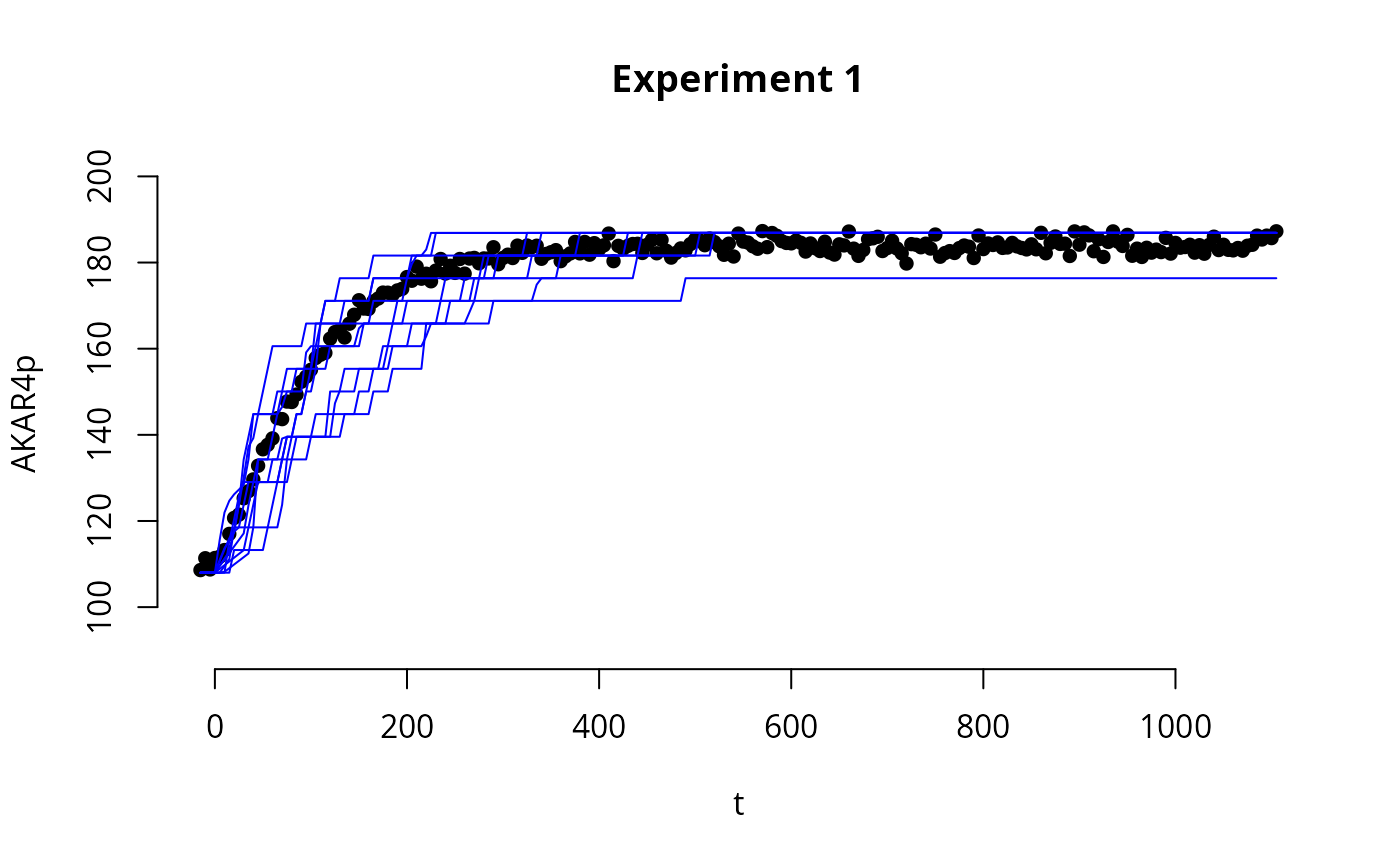
To check the fit of the sampled posterior parameters with the experimental data, we can simulate trajectories given sampled parameters and compare them with the experimental data.
expInd <- 1 # Index of the experiment that we want to plot, with corresponding fitted trajectories
e <- experiments[[1]]
# Plot the experimental data
par(bty='n',xaxp=c(80,120,4))
plot(e$outputTimes, e$outputValues$AKAR4pOUT,ylim=c(90,200), ylab="AKAR4p",
xlab="t",
main=sprintf("Experiment %i",expInd),
lwd=1.5,
pch=16)
# generate a function (s) that simulates a trajectory given a parameter in input
s <- simulator.stoch(experiments = experiments, model.tab = SBtab, parMap = parMap, outputFunction = model$func, vol = vol, unit = unit, nStochSim = 3)
n_draws <- dim(ABCMCMCoutput$draws)[1] # number of sampled posterior parameters
subsample_indices <- sample(n_draws,10) # indices of 10 random parameters out of the paramneters in
for(i in subsample_indices){
p <- ABCMCMCoutput$draws[i,] # random parameter from the posterior
y <- s(p) # simulate a trajectory (y) given parameter p
# "experiments" is a list of experiments
# "y" is a list where element i of the list is a simulation performed with the experimental conditions as in the i-th element of list "experiments"
lines(e$outputTimes, y[[expInd]]$output, col="blue") # plot the simulated trajectory
}
Predict unobserved compounds in the system
The data available for the AKA4 model consist of time series of concentrations of the compound AKAR4p. Using the posterior samples, we can predict the concentration of the unobserved compounds in the system. For example, here we show the predicted trajectories for the compound C.
expInd <- 1 # Index of the experiment that we want to plot, with corresponding fitted trajectories
e <- experiments[[1]]
# function that outputs the state C
output_AKAR4 <- function(t,state,param){state[1]}
# generate a function (s) that simulates a trajectory given a parameter in input
s <- simulator.stoch(experiments = experiments, model.tab = SBtab, parMap = parMap, outputFunction = output_AKAR4, vol = vol, unit = unit, nStochSim = 3)
n_draws <- dim(ABCMCMCoutput$draws)[1] # number of sampled posterior parameters
subsample_indices <- sample(n_draws,100) # indices of 10 random parameters out of the paramneters in
p <- ABCMCMCoutput$draws[subsample_indices[1],] # random parameter from the posterior
y <- s(p) # simulate a trajectory (y) given parameter p
# "experiments" is a list of experiments
# "y" is a list where element i of the list is a simulation performed with the experimental conditions as in the i-th element of list "experiments"
plot(e$outputTimes, y[[expInd]]$output, type="s", ylim=c(0,0.23), col=rgb(0,0,1,0.1), ylab = "AKAR4", xlab = "time") # plot the simulated trajectory
for(i in subsample_indices){
p <- ABCMCMCoutput$draws[i,] # random parameter from the posterior
y <- s(p) # simulate a trajectory (y) given parameter p
# "experiments" is a list of experiments
# "y" is a list where element i of the list is a simulation performed with the experimental conditions as in the i-th element of list "experiments"
lines(e$outputTimes, y[[expInd]]$output, type="s", col=rgb(0,0,1,0.1)) # plot the simulated trajectory
}