Importing Models into R
models.RmdIf the model can be described in terms of reactions, inputs, and outputs, then the most straightforward way to define it is through SBtab tables.1 2
The following R commands load the SBtab model and data into R as
data.frames, and convert the model into an ODE. First, you
will need to get to the directory with the SBtab files by the command
setwd(...).
f <- dir(pattern="[.]tsv$",full.names=TRUE) # file names
sb <- SBtabVFGEN::sbtab_from_tsv(f) # a list of data.frames
ml <- SBtabVFGEN::sbtab_to_vfgen(sb) # ODE model (as a list)
ex <- SBtabVFGEN::sbtab.data(sb,ml$conservationLaws) # includes the data
C <- uqsa::generateCode(ml) # C Code
cat(C, sep = "\n", file = "AKAR4cl_gvf.c")Some of the R commands above belong to our external package SBtabVFGEN.
More information on the R commands used in the previous code block, and further instructions on building and simulating your own model can be found in the pages “Build and simulate your own model” and “Importing Data”.
The rest of this page describes the variables and the files that we just created through the previous R commands.
Real Example AKAR4
library(uqsa)
library(SBtabVFGEN)
f <- uqsa_example('AKAR4') # file names
print(f[1])
sb <- SBtabVFGEN::sbtab_from_tsv(f) # a list of data.frames
ml <- SBtabVFGEN::sbtab_to_vfgen(sb) # model, as character vector
cl <- ml$conservationLaws # with conservation laws (if any)
ex <- SBtabVFGEN::sbtab.data(sb,cl) # list of simulation experimentsThe ex variable holds simulation instructions for the
different experiments as an R list that includes information on:
- inputs,
- initial values,
- measurement times,
- scheduled events,
- measured data.
SBtab contents include:
Reactions and conservation laws
print(sb$Reaction[,c(2,3)])
#> !KineticLaw !ReactionFormula
#> reaction_1 kf_C_AKAR4*C*AKAR4 - kb_C_AKAR4*AKAR4_C C + AKAR4 <=> AKAR4_C
#> reaction_2 kcat_AKARp*AKAR4_C AKAR4_C <=> AKAR4p + C
cat(cl$Text,sep="\n")
#> AKAR4_C_ConservedConst = AKAR4_C+1*C
#> AKAR4_ConservedConst = AKAR4+1*AKAR4p-1*CThe measured data, e.g. for experiment 1:
head(ex[[1]]$outputValues); cat("[...]\n"); tail(ex[[1]]$outputValues)
#> AKAR4pOUT
#> E400T001 108.60
#> E400T002 111.35
#> E400T003 108.75
#> E400T004 111.40
#> E400T005 111.70
#> E400T006 113.25
#> [...]
#> AKAR4pOUT
#> E400T220 184.125
#> E400T221 186.250
#> E400T222 185.350
#> E400T223 186.250
#> E400T224 185.675
#> E400T225 187.250The standard error of the data:
head(ex[[1]]$errorValues); cat("[...]\n"); tail(ex[[1]]$errorValues)
#> AKAR4pOUT
#> E400T001 14.7950
#> E400T002 14.9325
#> E400T003 14.8025
#> E400T004 14.9350
#> E400T005 14.9500
#> E400T006 15.0275
#> [...]
#> AKAR4pOUT
#> E400T220 18.57125
#> E400T221 18.67750
#> E400T222 18.63250
#> E400T223 18.67750
#> E400T224 18.64875
#> E400T225 18.72750Data as a plot:
i <- seq(70)
t <- ex[[1]]$outputTimes[i]
y <- ex[[1]]$outputValues[[1]][i]
dy <- ex[[1]]$errorValues[[1]][i]
if (require("errors")){
t <- set_errors(t,0)
y <- set_errors(y,dy)
plot(t,y)
} else {
plot(t,y,bty='n',ylim=c(100,200))
arrows(t,y,t,y+dy,angle=90, length=0.01)
arrows(t,y,t,y-dy,angle=90, length=0.01)
}
#> Loading required package: errors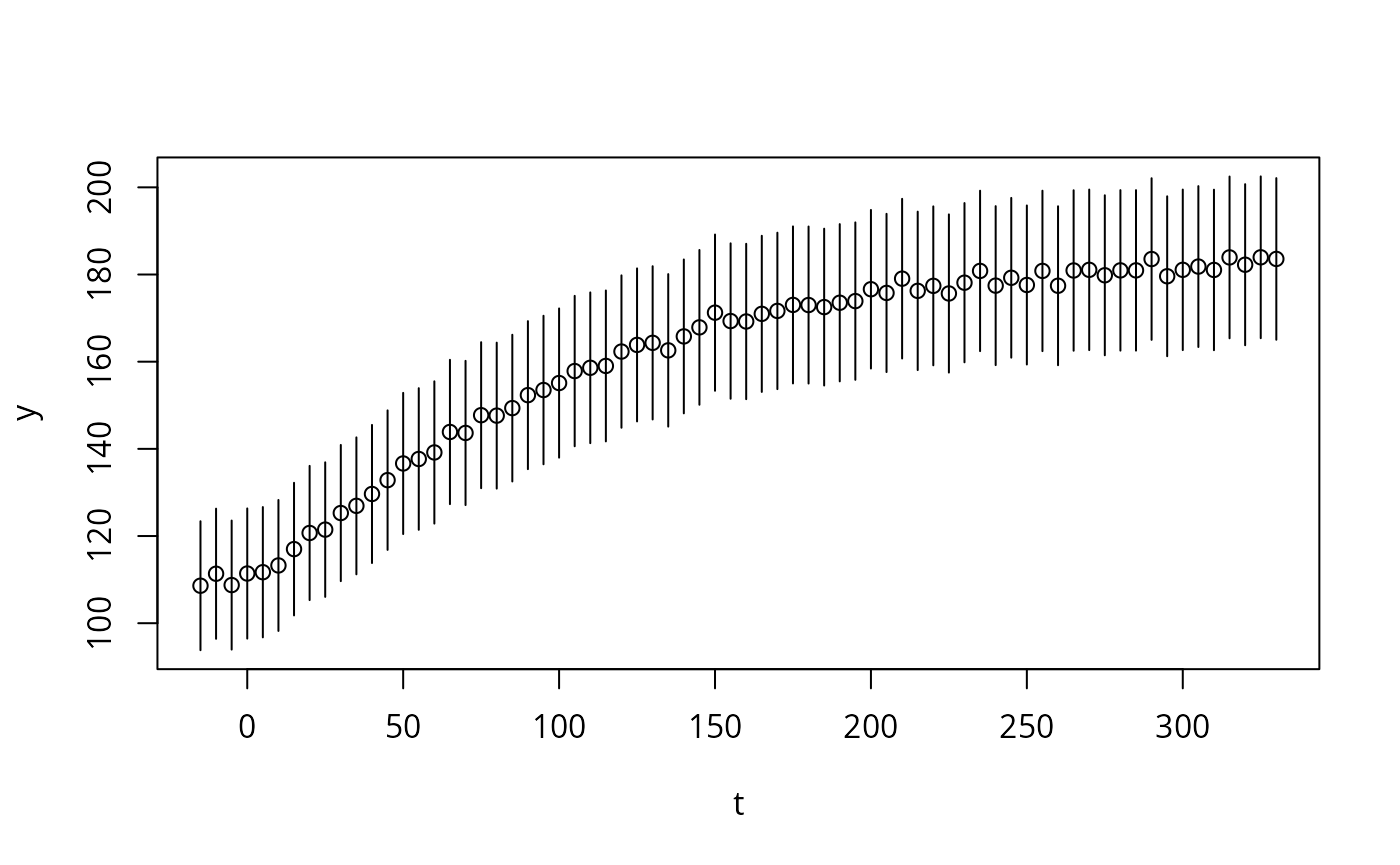
Files
The function sbtab_to_vfgen will produce several files,
with the model’s name (Document='model_name' attribute of
SBtab) and different extensions:
-
model_name.vf, a vfgen compatible file -
model_name.mod, a NEURON mod file - (optional)
model_name.xml, an SBML file, if you have the R bindings to libsbml installed in R and libsbml in your system -
model_name.tar.gz, compressed archive of a text representation of all ODE elements, no specific format -
model_name.zip, the same astar.gz, but as a zip archive
Let’s look at the state variables of the ode:
grep '<StateVariable' AKAR4.vf
#> <!-- <StateVariable Name="AKAR4" Description="removed compound" DefaultInitialCondition="0.2" Formula="-reaction_1"/> -->
#> <!-- <StateVariable Name="AKAR4_C" Description="removed compound" DefaultInitialCondition="0" Formula="+reaction_1-reaction_2"/> -->
#> <StateVariable Name="AKAR4p" Description="compound" DefaultInitialCondition="0" Formula="+reaction_2"/>
#> <StateVariable Name="C" Description="compound" DefaultInitialCondition="0" Formula="-reaction_1+reaction_2"/>Two State variables have been commented out (through conservation law analysis). These two have been replaced by vfgen expressions:
grep '<Expression' AKAR4.vf
#> <Expression Name="AKAR4_C" Description="derived from conservation law 1" Formula="AKAR4_C_ConservedConst - (C)"/>
#> <Expression Name="AKAR4" Description="derived from conservation law 2" Formula="AKAR4_ConservedConst - (AKAR4p-C)"/>
#> <Expression Name="reaction_1" Description="flux" Formula="kf_C_AKAR4*C*AKAR4 - kb_C_AKAR4*AKAR4_C"/>
#> <Expression Name="reaction_2" Description="flux" Formula="kcat_AKARp*AKAR4_C"/>Finally, we clean up the generated files:
for t in vf tar.gz zip ; do rm ./AKAR4.$t ; done
[ -f AKAR4.xml ] && rm AKAR4.xml # since that one is optional
echo $?The Vector Field file
The vf file is compatible with the vfgen tool, which
creates code in many languages from this vf file (MATLAB,
CVODE, Octave, Python, XPPAUT). Use this tool if you want to use the
created source code for other programming languages. Some SBtab content
will be missing here (e.g., scheduled events). The vfgen tool uses the
GiNaC library to calculate
Jacobians. GiNaC cannot parse vf files with inequality expressions in
them (e.g. a < b).
Converting the ODE to code
We use our tool, RPN-derivative, which
reads the same file (or one of the archive files:
zip/tar.gz) to create slightly different C
code. The main advantages are:
- a vector valued output function with a predictable name:
int MODEL_func() - the error code of each function is the length of the output buffers that need to be allocated
- more functions are created:
- initial conditions:
int MODEL_init() - default parameter values:
int MODEL_default()
- initial conditions:
The repository of this tool, icpm-kth/RPN-derivative, contains a
shell script ode.sh;
this script uses one of three methods to calculate analytical
derivatives for the Jacobians: 1. maxima, 2. yacas, 3. the
bin/derivative binary that RPN-derivative contains (after
make && make install)
Assuming that you followed all installation instructions:
Note: ode.sh is compatible with the same vf files from
SBtabVFGEN, but doesn’t support all of the output formats that VFGEN can
do, and does nothing related to delays.
Background on Parameters vs Inputs
Our goal is to perform parameter estimation. A systems biology model typically has parameters that are either reaction rate coefficients \(k_f, k_b\), equilibrium constants \(K_d\), Hill coefficients, and many other quantities related to kinetc laws. Parameters are often unknown or not uniquely determined yet. However, not all parameters of a model are unknown, and not all parameters are intrinsic to the system we study. Some of them describe the interventions we do to the system when performing an experiment. This could be the frequency of a driving force for a mechanical model, or the amount of a treatment dose (added to the system) that affects the time course of reactions:
- a substrate, or buffer
- an enzyme,
- a silencing agent,
- an inhibitor
These parameters are known to us, because they are written down in the protocol. If the input is time dependent, then its dynamics (or explicit algebraic functions) have to become part of the model. These input parameters can be different between experiments, but the intrinsic parameters (e.g., reaction rate coefficients) are always the same.
The ordinary differential equation, on the other hand, does not need to know the distinction between known or unknown parameters. For these reasons, we may write:
parMCMC <- [...] # some sampling variable
k <- parMap(parMCMC) # a model compatible parameter vector
u <- experiments[[i]]$input # a valid model input vector
p <- c(k,u)
# solve ODE using pwhere k are intrinsic, unknown parameters (of which we
quantify the uncertainty via sampling algorithms in UQSA), and
u are the known input parameters that are encoded in the
input field of an experiment.
If input parameters u exist, then they are always
concatenated (p <-c(k,u)) in that order (after
intrinsic, unknown parameters) and passed to the model. The model’s C
code only sees p. The wrapper functions in the
rgsl package do this concatenation when the solver is
called. We sample in logarithmic space, but the solver gets
k and appends the right u for each experiment,
both in linear space.
So, if parMCMC is in log10-space, then
parMap is the transformation to linear-space for the solver
to work in:
parMap <- function(par) {
return(10^(par))
}