Introduction
uqsa.RmdUQSA is an R-package for modelling and calibration of biochemical (and other) reaction networks.
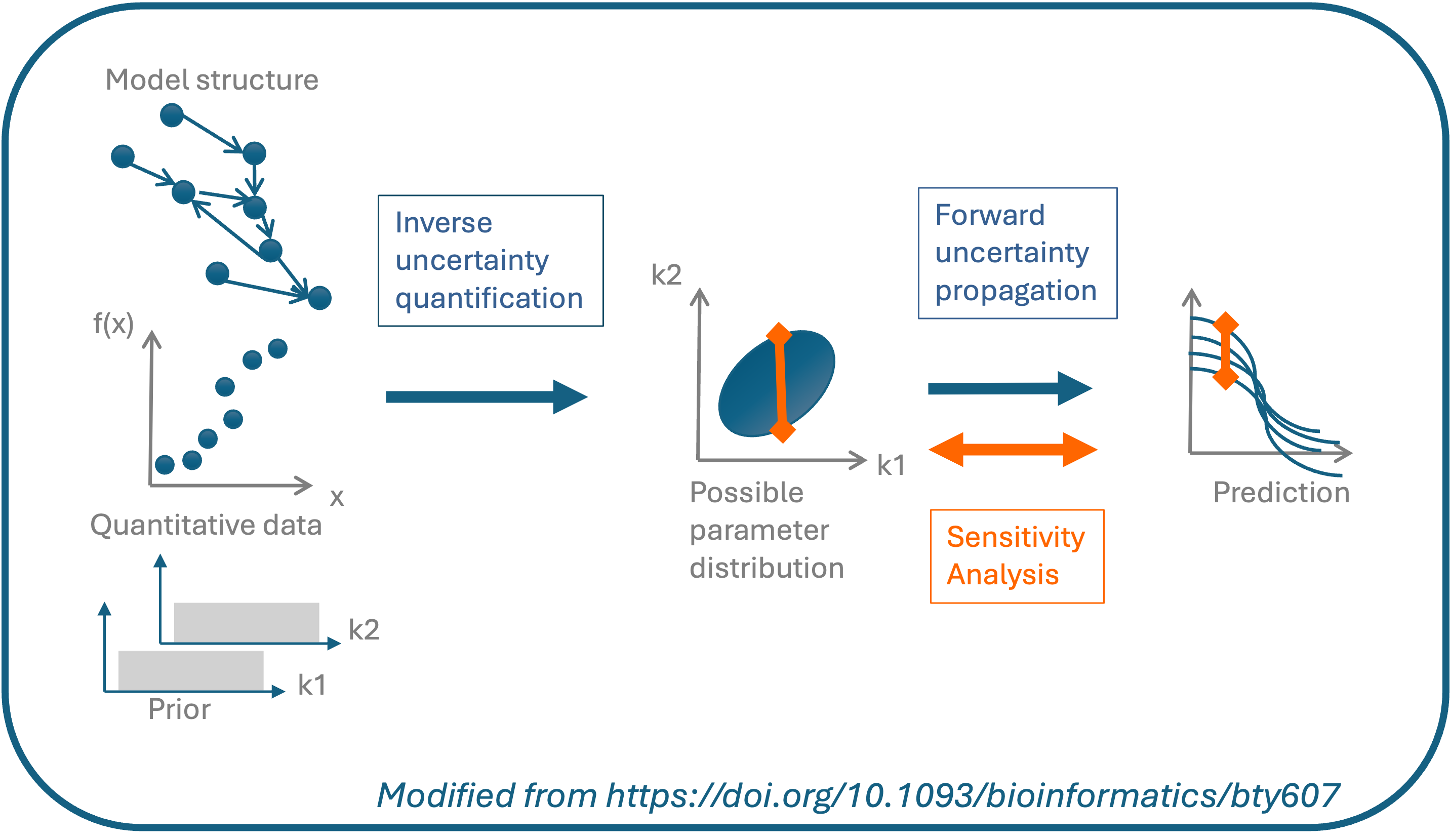
An important part of modelling is to estimate parameters
(calibration), and to account for the uncertainty in the parameter
estimates and predictions. UQSA allows you to perform Bayesian
uncertainty quantification while calibrating your model. With
UQSA you can also do a global sensitivity analysis to guide
further experiments.
For a quick hands-on introduction to UQSA, you can try our example on UQ and SA on the AKAR4 model.
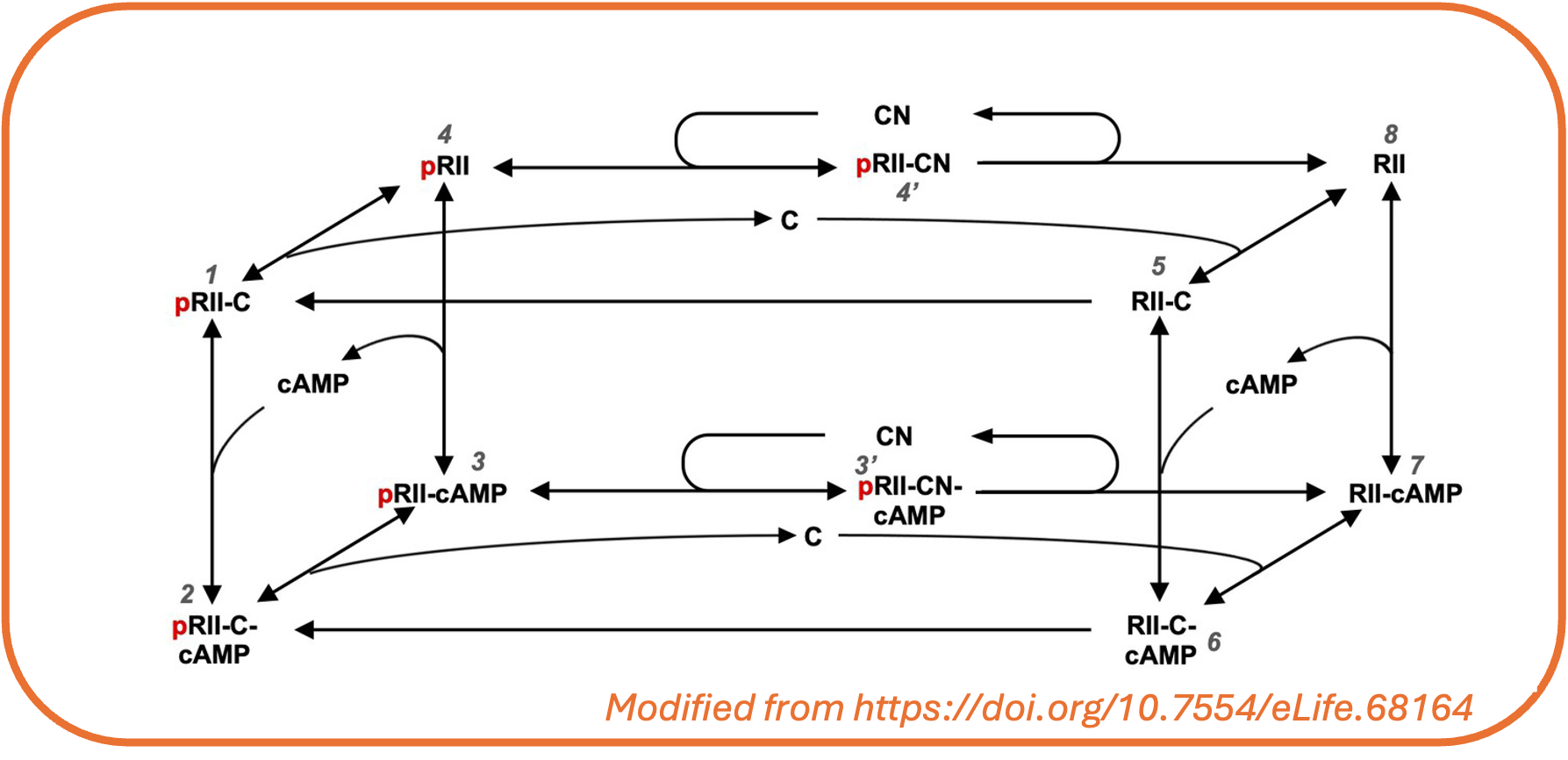
UQSA is specially constructed for systems biology models, as we use the SBtab format to define the model and calibration data (see intro to SBtab). SBtab files can be translated to and from other formats like SBML.
Models (reaction networks) written in SBtab can automatically be translated to ODE or stochastic models within UQSA. These mathematical models can next be simulated and calibrated. Uncertainty quantification is performed as part of the calibration. To do this, we use likelihood based Bayesian approaches for the ODE models and Approximate Bayesian Computation (ABC) for the stochastic models. Finally, a global sensitivity analysis can be performed on an independent or non-independent parameter space.
You can read more about UQSA in our article and explore several examples available in this documentation.