Global sensitivity analysis on AKAR4 - independent input factors
GSA_AKAR4.RmdThis article provides code do perform global sensitivity analysis with the Sobol-Saltelli method and the with the binning-approach.
Load the SBtab files, create ODE model code and load examples similar to the example Simulate the AKAR4 deterministic model.
modelFiles <- uqsa::uqsa_example("AKAR4")
SBtab <- SBtabVFGEN::sbtab_from_tsv(modelFiles)
model <- SBtabVFGEN::sbtab_to_vfgen(SBtab, cla=FALSE)
experiments <- SBtabVFGEN::sbtab.data(SBtab)
C <- uqsa::generateCode(model)
cat(C,sep="\n",file="./AKAR4_gvf.c")
modelName <- checkModel("AKAR4",modelFile="./AKAR4_gvf.c")Create simulator
We construct a simulator function and and test it on default parameter values.
s <- simcf(experiments,modelName,parMap=log10ParMap) # or simulator.c
p <- log10(SBtab$Parameter[["!DefaultValue"]])
names(p) <- rownames(SBtab$Parameter)
y <- s(p) # here the simulation happensSet meta-parameters for the global sensitivity simulation
nSamples <- 40000nSamples corresponds to the number of samples in M1 (or M2) of the Soboll-Saltelli approach..The number of simulations of the Sobol-Saltelli approach consists of 2*nSampels+nPars*nSamples number of simulations. In the binning approach below we use 2*nSamples number of samples (corresponding to 2*nSamples number of simulations) to use the same number of independent sample points as Sobol-Saltelli.
Construct parameter prior samples according to Sobol-Saltelli (M1, M2, N).
nPars <- length(p)
rprior <- rNormalPrior(p, array(1, nPars))
prior <- saltelli_prior(nSamples, rprior)
names(prior)
#> [1] "M1" "M2" "N"Plot parameter prior (M1)
title<-paste("Prior distribution experiment");
boxplot(prior$M1, main = title, names=names(p), las=2, cex.main=0.9, cex.axis=0.3)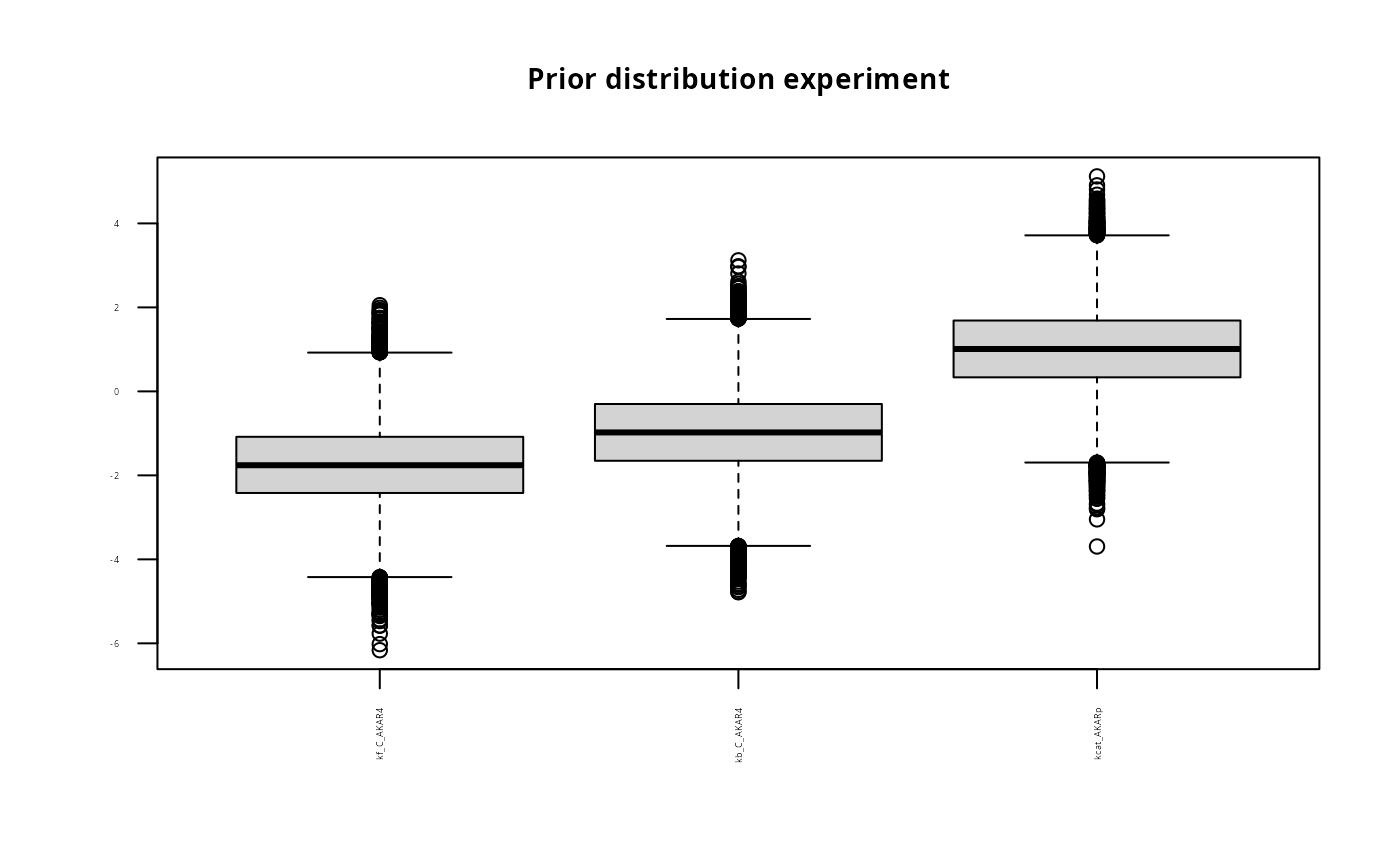
Simulate from the prior
Set up simulator considering one experiment
expIdx <- 2 #experiment to look at
s <- simcf(experiments[expIdx],modelName,parMap=log10ParMap)
#Test by simulting the default parameter set:
p <- log10(SBtab$Parameter[["!DefaultValue"]])
names(p) <- rownames(SBtab$Parameter)
y <- s(p) # here the simulation happensUse states (compound concentrations) as output and look at one time point alone
T <-5 #timepoint to investigate
nStates <- dim(y[[1]]$state)[1]
fM1 <- t(s(t(prior$M1))[[1]]$state[,T,])
fM2 <- t(s(t(prior$M2))[[1]]$state[,T,])
fN <- array(NA, dim=c(nSamples,nStates, nPars))
for (i in 1:nPars){
print(i)
fN[,,i] <- t(s(t(prior$N[,,i]))[[1]]$state[,T,])
}
#> [1] 1
#> [1] 2
#> [1] 3Calculate and plot sensitivity indexes
Calculate sensitivity indexes for sobol-saltelli
SA <- gsa_saltelli(fM1,fM2,fN)Plot first (SI) and total-order (SIT) sensitivity indexes for sobol-saltelli
cols=rainbow(3)
par(mfrow = c(1, 2))
barplot(t(SA$SI[,]),
col=cols,
border="white",
space=0.04,
cex.axis=1,
names.arg=dimnames(y[[1]]$state)[[1]],
cex.names = 0.7,
las = 2,
xlab="SI", las=2, cex.main=0.9)
barplot(t(SA$SIT[,]),
col=cols,
border="white",
space=0.04,
cex.axis=1,
names.arg=dimnames(y[[1]]$state)[[1]],
cex.names = 0.7,
las = 2,
xlab="SIT",
legend.text=SBtab$Parameter[,1],
args.legend = list(x = "topright", inset=c(0, 0.1), cex=0.7))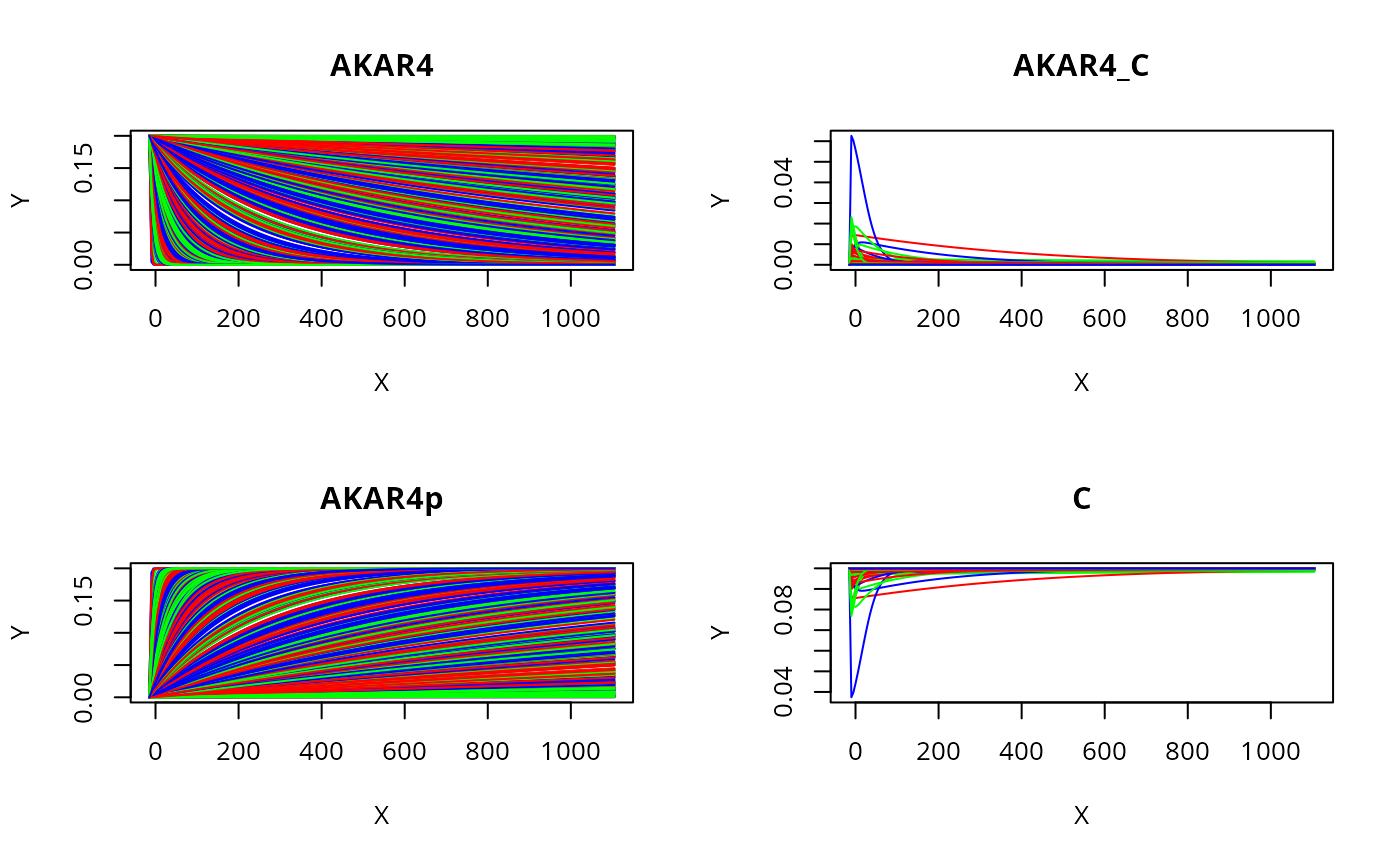
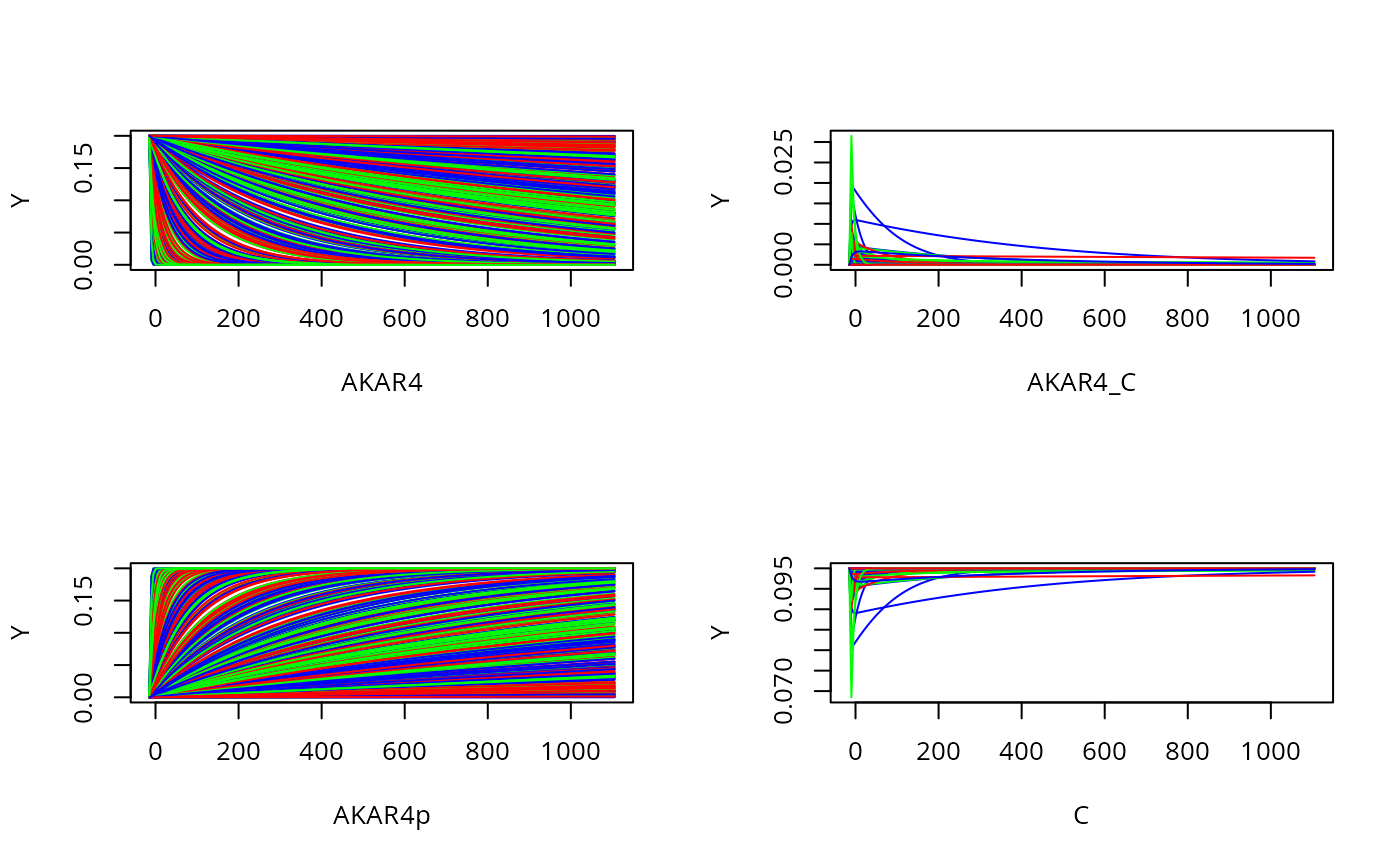
Plot all states and timeponts for the experiment
allTimesSample=s(t(prior$M2))[[1]]$state
par(mfrow = c(2, 2))
for (i in 1:4) {
matplot(experiments[[1]]$outputTime,allTimesSample[i,,1:500] , type = "l", lty = 1,
col = c("red", "blue", "green"),
ylab = "Y", xlab= dimnames(allTimesSample)[[1]][i])
}
Calculate first order sensitivity index (SI) based on binning approach
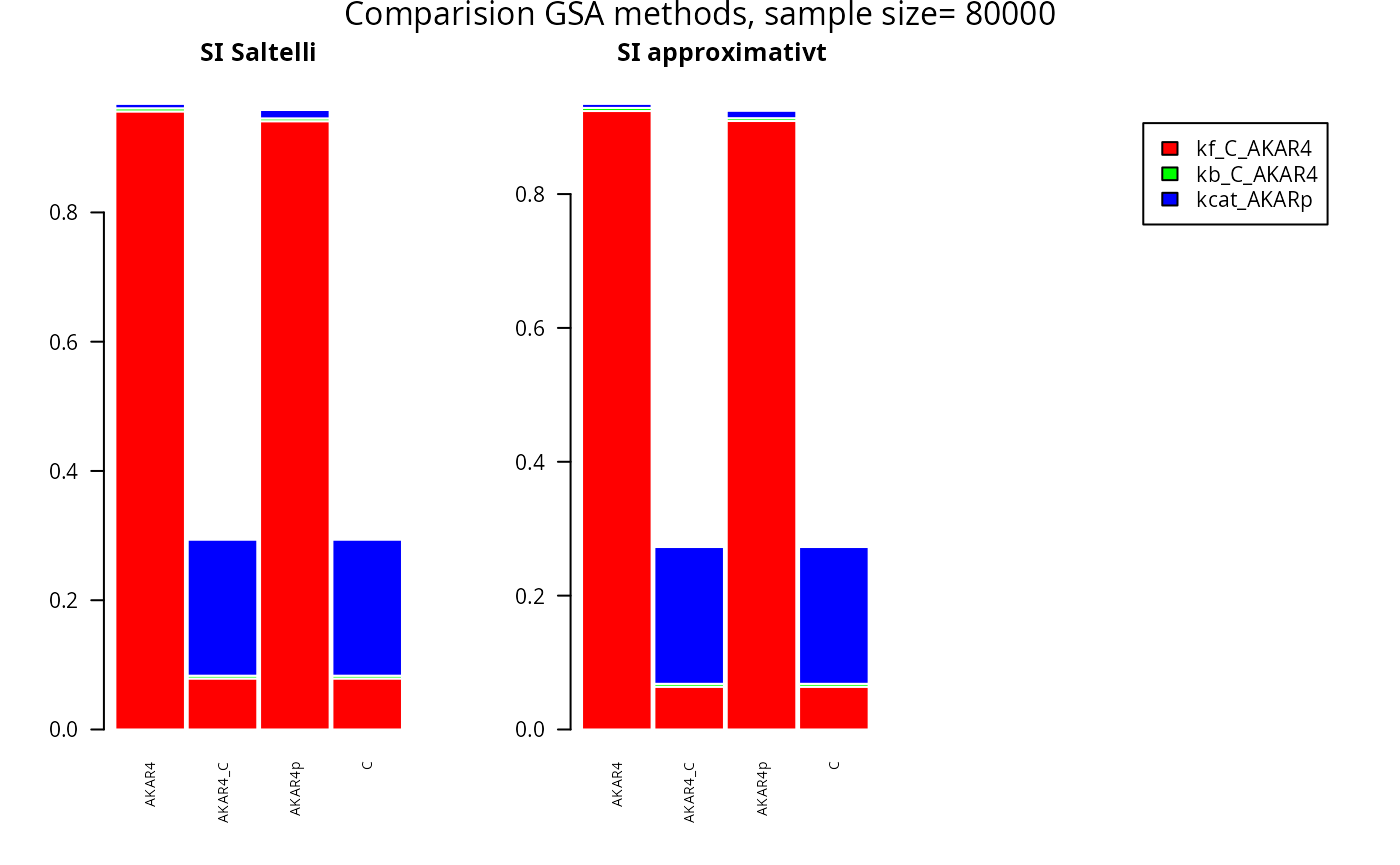
SIappr <-gsa_binning(rbind(prior$M1,prior$M2), rbind(fM1,fM2), nBins = "Sturges")Plot SI for Sobol-Saltelli versus the binning approach
par(mfrow = c(1, 3))
barplot(t(SA$SI[,]),
col=cols,
border="white",
space=0.04,
cex.axis=1,
names.arg=dimnames(y[[1]]$state)[[1]],
cex.names = 0.7,
las = 2,
main="SI Saltelli")
barplot(t(SIappr),
col=cols,
border="white",
space=0.04,
cex.axis=1,
cex.names = 0.7,
las = 2,
main="SI approximativt ")
barplot(c(0),
axes=FALSE,
col=cols,
border="white",
space=0.04,
font.axis=2,
legend.text=SBtab$Parameter[,1])
mtext(paste("Comparision GSA methods, sample size=",as.character(2*nSamples)), side = 3, line = -1.2, outer = TRUE)