Transformation Events
events.RmdThe solver we use in most of our examples is
rgsl::r_gsl_odeiv2_outer which has the built-in ability to
perform sudden interventions. Here we show an example of how this can be
used to trigger sudden spikes (action potentials)
f <- uqsa_example('Spike')
sb <- sbtab_from_tsv(f)
#> [tsv] file[1] «AP20Hz.tsv» belongs to Document «Spike»
#> I'll take this as the Model Name.
#> AP20Hz.tsv Compound.tsv E20Hz.tsv Experiment.tsv Input.tsv Output.tsv Parameter.tsv Reaction.tsv Transformation.tsv
ex <- sbtab.data(sb)The transformations are still present in sb:
sb$Transformation
#> >Ca >Buffer
#> APCa Ca A*(k2-k1)*doseBut, also built into the model and simulation instructions
ex
ex[[1]]$events
#> $time
#> [1] 0 50 100 150 200 250 300 350 400
#>
#> $label
#> [1] 0 0 0 0 0 0 0 0 0
#>
#> $dose
#> [1] 1 1 1 1 1 1 1 1 1Simulation
cfile <- uqsa_example("Spike",pat='_gvf[.]c$')
modelName <- checkModel("Spike",cfile)
#> building a shared library from c source, and using GSL odeiv2 as backend (pkg-config is used here).
#> cc -shared -fPIC `pkg-config --cflags gsl` -o './Spike.so' '/tmp/Rtmp3XbkUp/temp_libpath315eec67fa214d/uqsa/extdata/Spike/Spike_gvf.c' `pkg-config --libs gsl`
t <- seq(-4,1000)
ex[[1]]$outputTimes <- t
SIM <- simcf(ex,modelName)
file.exists(comment(modelName))
#> [1] TRUE
par <- sb$Parameter[["!DefaultValue"]]
print(par)
#> [1] 9.78422e-03 3.44800e-02 2.29127e+02
res <- SIM(par)
y <- as.numeric(res[[1]]$func)
plot(t,y,bty="n",type='l',xlab="t",ylab="Ca")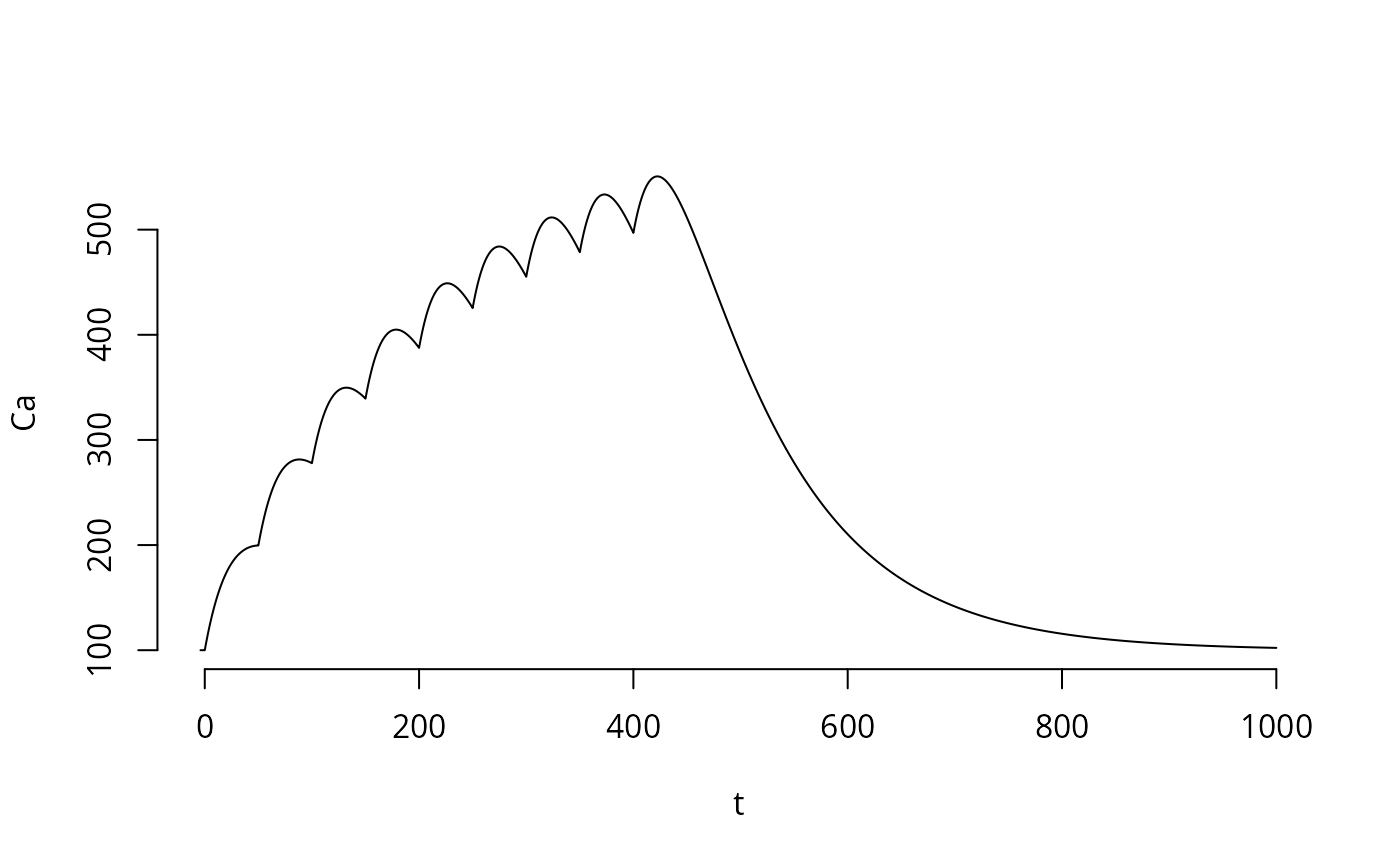
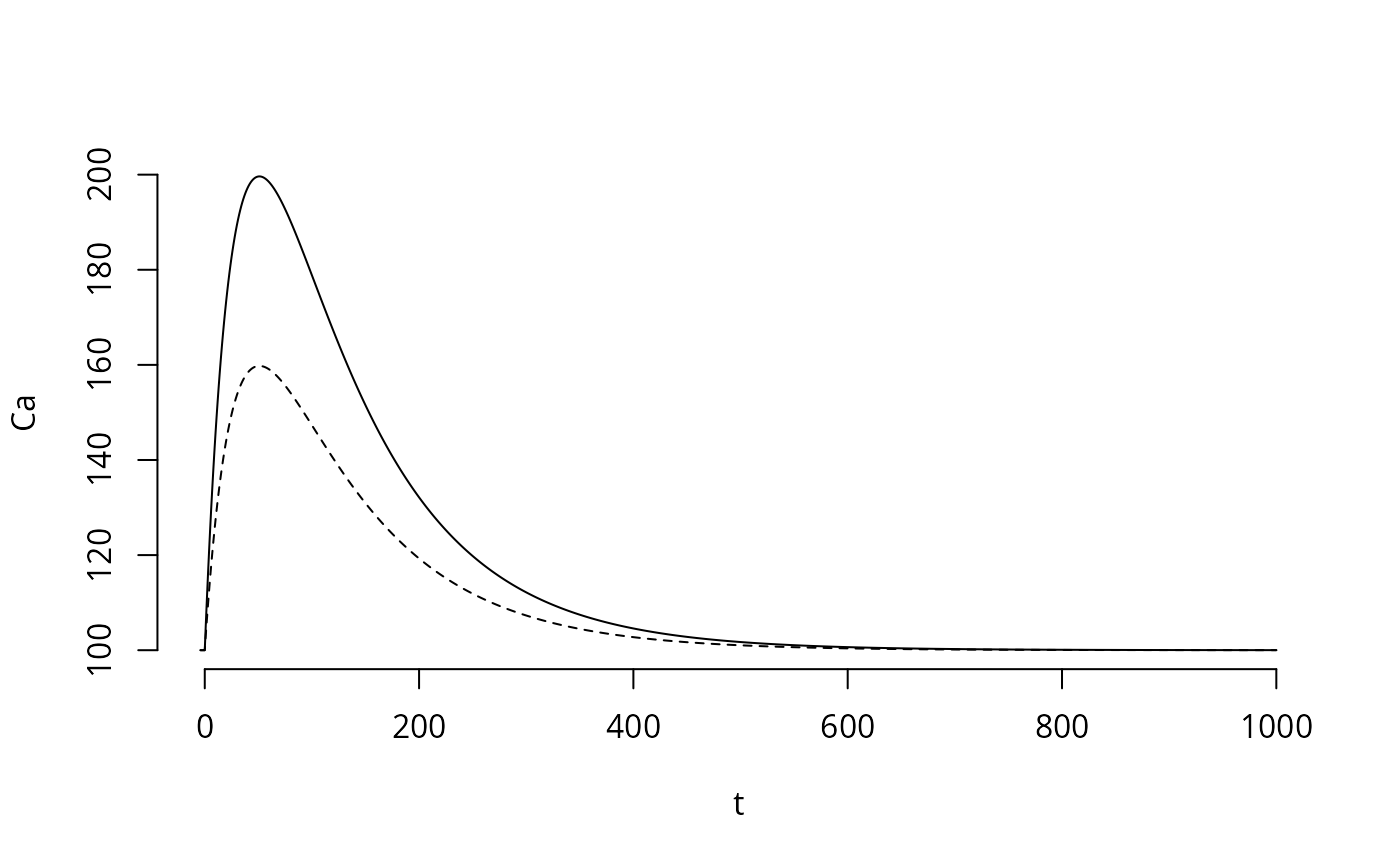
Maybe, we should also simulate how one Ca spike would look like, and also try reducing the dose a bit:
ex[[1]]$event$time <- 0.0
ex[[1]]$event$label <- 0
ex[[1]]$event$dose <- 1.0
ex[[2]] <- ex[[1]]
ex[[2]]$event$dose <- 0.6
SIM <- simcf(ex,modelName)
par <- sb$Parameter[["!DefaultValue"]]
print(par)
#> [1] 9.78422e-03 3.44800e-02 2.29127e+02
res <- SIM(par)
y1 <- as.numeric(res[[1]]$func)
y2 <- as.numeric(res[[2]]$func)
plot(t,y1,bty="n",type='l',xlab="t",ylab="Ca")
lines(t,y2,lty=2)